2025
Yi, L., Zhou, Y., Zhang, C., Lu, H., & Xie, Y. (2025),
GlycoRNA research: from unknown unknowns to known unknowns.
Protein & Cell. Advance online publication.
Porat, J., Poli, V., Almahayni, K., George, B. M., de Luna Vitorino, F. N., Yi, L., Costa, C. B., Zhou, Y., Crompton, A., Wen, H., Zheng, L., Gregersen, P. K., Agarwal, S., Garcia, B. A., Xie, Y., Möckl, L., Zanoni, I., Simpfendorfer, K. R., & Flynn, R. A.(2025),
DNASE1L3 surveils mitochondrial DNA on the surface of distinct mammalian cells.
bioRxiv, 2025.10.05.680464.
Lin, Z., Xie, Y., Gongora, J., Liu, X., Zahn, E., Palai, B. B., Ramirez, D. H., Searfoss, R. M., Vitorino, F. N., Karki, R., Dann, G. P., Zhao, C., Han, X., MacTaggart, B., Lan, X., Fu, D., Greenberg, L., Zhang, Y., Lavine, K. J., Greenberg, M. J., … Garcia, B. A. (2025),
An unbiased proteomic platform for ATE1-based arginylation profiling..
Nature chemical biology, 10.1038/s41589-025-01996-z. Advance online publication.
Zeng, G.-D., Wang, Y.-L., Chen, X.-H., Ma, Y., Zhu, Z., Liu, H.-L., Fan, L.-Y., Nan, H., Nur, Z., Wei, M., Liu, X.-Y., Yi, L., Jin, J.-Y., Cao, Y.-S., Qiu, M.-B., Sun, W.-P., Bai, Y., Zhou, Q.-B., Zhang, G.-Q., Chen, J.-F., Shi, J., Zheng, J., Xie, Y., Li, P., Wan, X.-B., & Fan, X.-J.(2025),
StCEL-based platform enables detection and sequencing of native sialoglycoRNA.
bioRxiv, 2025.08.06.668864.
Liu, X., Yi, L., Lin, Z., Chen, S., Wang, S., Sheng, Y., Lebrilla, C. B., Garcia, B. A., & Xie, Y. (2024),
Metabolic control of glycosylation forms for establishing glycan-dependent protein interaction networks.
Proceedings of the National Academy of Sciences of the United States of America, vol. 122,25 (2025): e2422936122.
Chen, S., Xie, Y., Alvarez, M. R., Sheng, Y., Bouchibti, Y., Chang, V., & Lebrilla, C. B. (2025).
Quantitative Glycan-Protein Cross-Linking Mass Spectrometry Using Enrichable Linkers Reveals Extensive Glycan-Mediated Protein Interaction Networks.
Analytical chemistry, 10.1021/acs.analchem.4c04134.
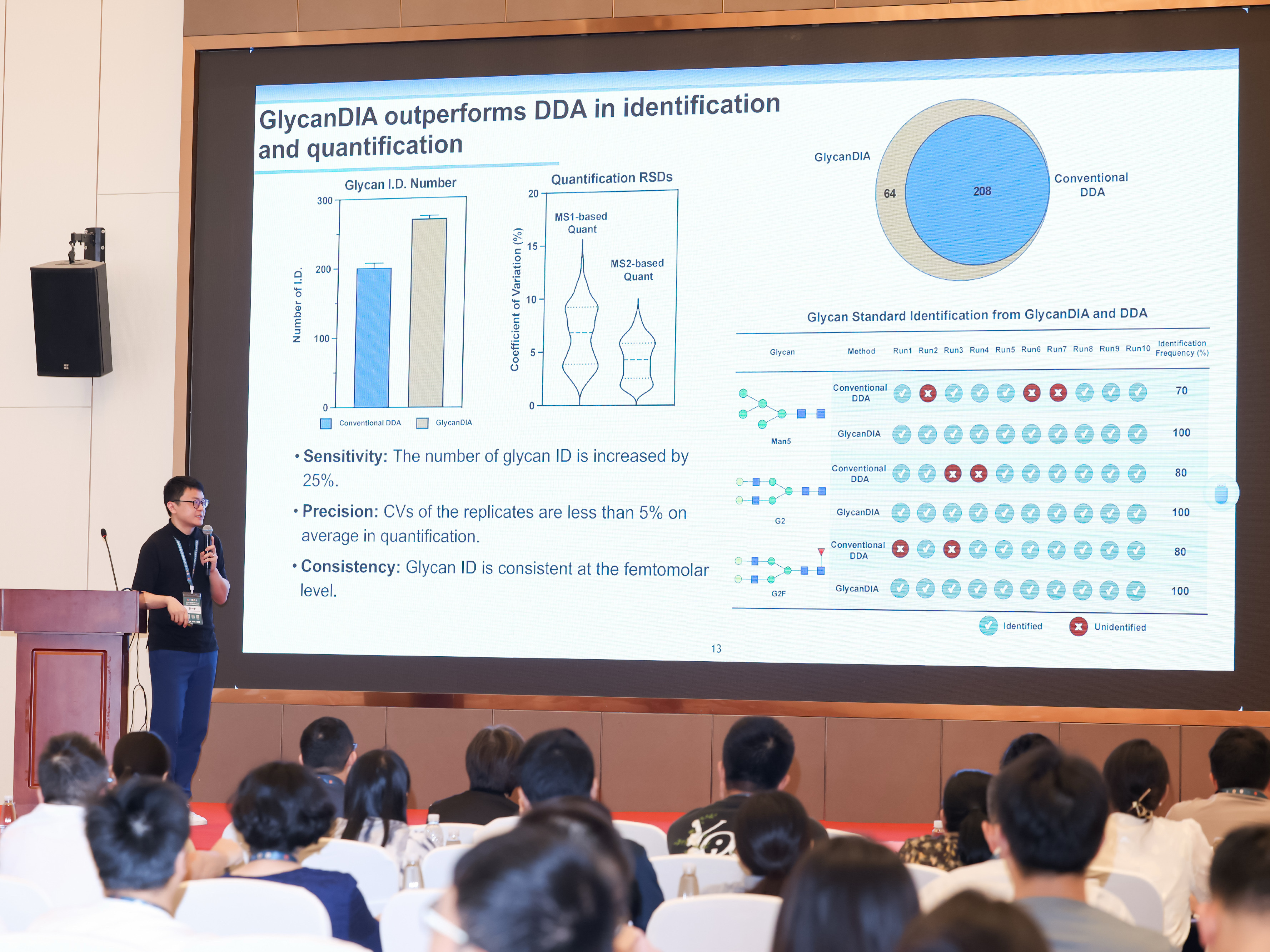
Xie, Y., Liu, X., Yi, L., Wang, S., Lin, Z., Zhao, C., Chen, S., Robison, F. M., George, B. M., Lebrilla, C. B., Flynn, R. A., & Garcia, B. A. (2025).
Development and application of GlycanDIA workflow for glycomic analysis.
Nature Communications, 16(1), 7075.
Liu, X., Yi, L., Lin, Z., Chen, S., Wang, S., Sheng, Y., Lebrilla, C. B., Garcia, B. A., & Xie, Y. (2024),
Metabolic control of glycosylation forms for establishing glycan-dependent protein interaction networks.
Proceedings of the National Academy of Sciences of the United States of America vol. 122,25 (2025): e2422936122.
Prior to Independent Research
Xie, Y., Chai, P., Till, N. A., Hemberger, H., Lebedenko, C. G., Porat, J., Watkins, C. P., Caldwell, R. M., George, B. M., Perr, J., Bertozzi, C. R., Garcia, B. A., & Flynn, R. A. (2024), The modified RNA base acp3U is an attachment site for N-glycans in glycoRNA. Cell, 187(19), 5228–5237.e12.
Porat, J., Watkins, C. P., Jin, C., Xie, Y., Tan, X., Lebedenko, C. G., Hemberger, H., Shin, W., Chai, P., Collins, J. J., Garcia, B. A., Bojar, D., & Flynn, R. A. (2024), O-glycosylation contributes to mammalian glycoRNA biogenesis. bioRxiv: the preprint server for biology, 2024.08.28.610074.
Liu, X., Zhang, Y., Wen, Z., Hao, Y., Banks, C. A. S., Cesare, J., Bhattacharya, S., Arvindekar, S., Lange, J. J., Xie, Y., Garcia, B. A., Slaughter, B. D., Unruh, J. R., Viswanath, S., Florens, L., Workman, J. L., & Washburn, M. P. (2024), An integrated structural model of the DNA damage-responsive H3K4me3 binding WDR76:SPIN1 complex with the nucleosome. Proceedings of the National Academy of Sciences of the United States of America, 121(33), e2318601121.
Lin, Z., Xie, Y., Gongora, J., Liu, X., Zahn, E., Palai, B. B., Ramirez, D., Searfoss, R. M., Vitorino, F. N., Dann, G. P., Zhao, C., Han, X., MacTaggart, B., Lan, X., Fu, D., Greenberg, L., Zhang, Y., Lavine, K. J., Greenberg, M. J., Lv, D., … Garcia, B. A. (2024), An unbiased proteomic platform for activity-based arginylation profiling. bioRxiv : the preprint server for biology, 2024.06.01.596974.
Izadifar, Z., Cotton, J., Chen, S., Horvath, V., Stejskalova, A., Gulati, A., LoGrande, N. T., Budnik, B., Shahriar, S., Doherty, E. R., Xie, Y., To, T., Gilpin, S. E., Sesay, A. M., Goyal, G., Lebrilla, C. B., & Ingber, D. E. (2024), Mucus production, host-microbiome interactions, hormone sensitivity, and innate immune responses modeled in human cervix chips. Nature Communications, 15(1), 4578.
Xie, Y., Chen, S., Alvarez, M. R., Sheng, Y., Li, Q., Maverakis, E., & Lebrilla, C. B. (2024),
Protein oxidation of fucose environments (POFE) reveals fucose-protein interactions.
Chemical Science, 15(14), 5256–5267.
Frisbie, V. S., Hashimoto, H., Xie, Y., De Luna Vitorino, F. N., Baeza, J., Nguyen, T., Yuan, Z., Kiselar, J., Garcia, B. A., & Debler, E. W. (2024),
Two DOT1 enzymes cooperatively mediate efficient ubiquitin-independent histone H3 lysine 76 tri-methylation in kinetoplastids.
Nature Communications, 15(1), 2467.
Lin, Z., Gongora, J., Liu, X., Xie, Y., Zhao, C., Lv, D., & Garcia, B. A. (2023),
Automation to enable high-throughput chemical proteomics.
Journal of Proteome Research, 22(12), 3676–3682.
Lu-Culligan, W. J., Connor, L. J., Xie, Y., Ekundayo, B. E., Rose, B. T., Machyna, M., Pintado-Urbanc, A. P., Zimmer, J. T., Vock, I. W., Bhanu, N. V., King, M. C., Garcia, B. A., Bleichert, F., & Simon, M. D. (2023),
Acetyl-methyllysine marks chromatin at active transcription start sites.
Nature, 622(7981), 173–179.
Chen, S., Bouchibti, Y., Xie, Y., Chen, Y., Chang, V., & Lebrilla, C. B. (2023),
Analysis of cell glycogen with quantitation and determination of branching using liquid chromatography-mass spectrometry.
Analytical Chemistry, 95(34), 12884–12892.
Lauman, R., Kim, H. J., Pino, L. K., Scacchetti, A., Xie, Y., Robison, F., Sidoli, S., Bonasio, R., & Garcia, B. A. (2023). Expanding the epitranscriptomic RNA sequencing and modification mapping mass spectrometry toolbox with field asymmetric waveform ion mobility and electrochemical elution liquid chromatography. Analytical Chemistry, 95(12), 5187–5195.
Alvarez, M. R., Zhou, Q., Tena, J., Barboza, M., Wong, M., Xie, Y., Lebrilla, C. B., Cabanatan, M., Barzaga, M. T., Tan-Liu, N., Heralde, F. M., 3rd, Serrano, L., Nacario, R. C., & Completo, G. C. (2023). Glycomic, glycoproteomic, and proteomic profiling of philippine lung cancer and peritumoral tissues: case series study of patients stages I–III. Cancers, 15(5), 1559.
Xie, Y., De Luna Vitorino, F. N., Chen, Y., Lempiäinen, J. K., Zhao, C., Steinbock, R. T., Lin, Z., Liu, X., Zahn, E., Garcia, A. L., Weitzman, M. D., & Garcia, B. A. (2023). SWAMNA: a comprehensive platform for analysis of nucleic acid modifications. Chemical Communications (Cambridge, England), 59(83), 12499–12502.
Merleev, A. A., Le, S. T., Alexanian, C., Toussi, A., Xie, Y., Marusina, A. I., Watkins, S. M., Patel, F., Billi, A. C., Wiedemann, J., Izumiya, Y., Kumar, A., Uppala, R., Kahlenberg, J. M., Liu, F. T., Adamopoulos, I. E., Wang, E. A., Ma, C., Cheng, M. Y., Xiong, H., … Maverakis, E. (2022), Biogeographic and disease-specific alterations in epidermal lipid composition and single-cell analysis of acral keratinocytes. JCI Insight, 7(16), e159762.
Xie, Y., Janssen, K. A., Scacchetti, A., Porter, E. G., Lin, Z., Bonasio, R., & Garcia, B. A. (2022), Permethylation of ribonucleosides provides enhanced mass spectrometry quantification of post-transcriptional RNA modifications. Analytical Chemistry, 94(20), 7246–7254.
Janssen, K. A., Xie, Y., Kramer, M. C., Gregory, B. D., & Garcia, B. A. (2022), Data-independent acquisition for the detection of mononucleoside RNA modifications by mass spectrometry. Journal of the American Society for Mass Spectrometry, 33(5), 885–893.
Sheng, Y., Vinjamuri, A., Alvarez, M. R. S., Xie, Y., McGrath, M., Chen, S., Barboza, M., Frieman, M., & Lebrilla, C. B. (2022), Host cell glycocalyx remodeling reveals SARS-CoV-2 Spike protein glycomic binding sites. Frontiers in Molecular Biosciences, 9, 799703.
Li, Q., Xie, Y., Rice, R., Maverakis, E., & Lebrilla, C. B. (2022), A proximity labeling method for protein-protein interactions on cell membrane. Chemical Science, 13(20), 6028–6038.
Kim, T., Xie, Y., Li, Q., Artegoitia, V. M., Lebrilla, C. B., Keim, N. L., Adams, S. H., & Krishnan, S. (2021), Diet affects glycosylation of serum proteins in women at risk for cardiometabolic disease. European Journal of Nutrition, 60(7), 3727–3741.
Zhou, Q., Xie, Y., Lam, M., & Lebrilla, C. B. (2021), N-Glycomic analysis of the cell shows specific effects of glycosyl transferase inhibitors. Cells, 10(9), 2318.
Maverakis, E., Merleev, A. A., Park, D., Kailemia, M. J., Xu, G., Ruhaak, L. R., Kim, K., Hong, Q., Li, Q., Leung, P., Liakos, W., Wan, Y. Y., Bowlus, C. L., Marusina, A. I., Lal, N. N., Xie, Y., Luxardi, G., & Lebrilla, C. B. (2021), Glycan biomarkers of autoimmunity and bile acid-associated alterations of the human glycome: primary biliary cirrhosis and primary sclerosing cholangitis-specific glycans. Clinical Immunology, 230, 108825.
Zheng, J. J., Agus, J. K., Hong, B. V., Tang, X., Rhodes, C. H., Houts, H. E., Zhu, C., Kang, J. W., Wong, M., Xie, Y., Lebrilla, C. B., Mallick, E., Witwer, K. W., & Zivkovic, A. M. (2021), Isolation of HDL by sequential flotation ultracentrifugation followed by size exclusion chromatography reveals size-based enrichment of HDL-associated proteins. Scientific Reports, 11(1), 16086.
Xie, Y., Chen, S., Li, Q., Sheng, Y., Alvarez, M. R., Reyes, J., Xu, G., Solakyildirim, K., & Lebrilla, C. B. (2021), Glycan-protein cross-linking mass spectrometry reveals sialic acid-mediated protein networks on cell surfaces. Chemical Science, 12(25), 8767–8777.
Kasper, D. M., Hintzen, J., Wu, Y., Ghersi, J. J., Mandl, H. K., Salinas, K. E., Armero, W., He, Z., Sheng, Y., Xie, Y., Heindel, D. W., Park, E. J., Sessa, W. C., Mahal, L. K., Lebrilla, C., Hirschi, K. K., & Nicoli, S. (2020), The N-glycome regulates the endothelial-to-hematopoietic transition. Science, 370(6521), 1186–1191.
Merleev, A. A., Park, D., Xie, Y., Kailemia, M. J., Xu, G., Ruhaak, L. R., Kim, K., Hong, Q., Li, Q., Patel, F., Wan, Y. Y., Marusina, A. I., Adamopoulos, I. E., Lal, N. N., Mitra, A., Le, S. T., Shimoda, M., Luxardi, G., Lebrilla, C. B., & Maverakis, E. (2020), A site-specific map of the human plasma glycome and its age and gender-associated alterations. Scientific Reports, 10(1), 17505.
Park, S., Doherty, E. E., Xie, Y., Padyana, A. K., Fang, F., Zhang, Y., Karki, A., Lebrilla, C. B., Siegel, J. B., & Beal, P. A. (2020), High-throughput mutagenesis reveals unique structural features of human ADAR1. Nature Communications, 11(1), 5130.
Li, Q., Xie, Y., Wong, M., Barboza, M., & Lebrilla, C. B. (2020), Comprehensive structural glycomic characterization of the glycocalyxes of cells and tissues. Nature Protocols, 15(8), 2668–2704.
Xie, Y., Sheng, Y., Li, Q., Ju, S., Reyes, J., & Lebrilla, C. B. (2020), Determination of the glycoprotein specificity of lectins on cell membranes through oxidative proteomics. Chemical Science, 11(35), 9501–9512.
Xie, Y., Peng, C., Gao, Y., Liu, X., Liu, T., & Joy, A. (2017). Mannose-based graft polyesters with tunable binding affinity to concanavalin A. Journal of Polymer Science Part A: Polymer Chemistry, 55, 3908-3917.